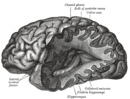
Neuroanatomy
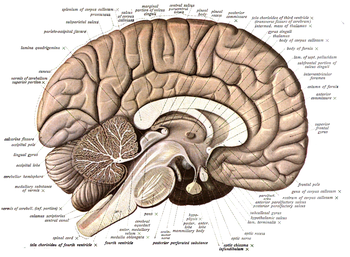
Neuroanatomy is the study of the anatomy and organisation of the nervous system. Pictured here is a cross-section showing the gross anatomy of the human brain
Neuroanatomy is the study of the structure and organization of the nervous system. In contrast to animals with radial symmetry, whose nervous system consists of a distributed network of cells, animals with bilateral symmetry have segregated, defined nervous systems. Their neuroanatomy is therefore better understood. In vertebrates, the nervous system is segregated into the internal structure of the brain and spinal cord (together called the central nervous system, or CNS) and the routes of the nerves that connect to the rest of the body (known as the peripheral nervous system, or PNS). The delineation of distinct structures and regions of the nervous system has been critical in investigating how it works. For example, much of what neuroscientists have learned comes from observing how damage or "lesions" to specific brain areas affects behavior or other neural functions.
For information about the composition of non-human animal nervous systems, see nervous system. For information about the typical structure of the Homo sapiens nervous system, see human brain or peripheral nervous system. This article discusses information pertinent to the study of neuroanatomy.
Contents
1 History
2 Composition
2.1 Orientation in neuroanatomy
3 Tools
3.1 Cell staining
3.2 Histochemistry
3.3 Genetically encoded markers
3.4 Non-invasive brain imaging
3.5 Viral-based methods
3.6 Dye-based methods
3.7 Connectomics
3.8 Computational neuroanatomy
4 Model systems
4.1 Caenorhabditis elegans
4.2 Drosophila melanogaster
5 See also
6 References
7 External links
History

J. M. Bourgery's anatomy of the brain, brainstem, and upper spinal column
The first known written record of a study of the anatomy of the human brain is the ancient Egyptian document the Edwin Smith Papyrus.[1] The next major development in neuroanatomy came from the Greek Alcmaeon, who determined that the brain and not the heart ruled the body and that the senses were dependent on the brain.[2]
After Alcmaeon’s findings, many scientists, philosophers, and physicians from around the world continued to contribute to the understanding of neuroanatomy, notably: Galen, Herophilus, Rhazes and Erasistratus. Herophilus and Erasistratus of Alexandria were perhaps the most influential Greek neuroscientists with their studies involving dissecting the brains.[2] For several hundred years afterward, with the cultural taboo of dissection, no major progress occurred in neuroscience. However, Pope Sixtus IV effectively revitalized the study of neuroanatomy by altering the papal policy and allowing human dissection. This resulted in a boom of research in neuroanatomy by artists and scientists of the Renaissance.[3]
In 1664, Thomas Willis, a physician and professor at Oxford University, coined the term neurology when he published his text Cerebri anatome which is considered the foundation of neuroanatomy.[4] The subsequent three hundred and fifty some years has produced a great deal of documentation and study of the neural systems.
Composition
At the tissue level, the nervous system is composed of neurons, glial cells, and extracellular matrix. Both neurons and glial cells come in many types (see, for example, the nervous system section of the list of distinct cell types in the adult human body). Neurons are the information-processing cells of the nervous system: they sense our environment, communicate with each other via electrical signals and chemicals called neurotransmitters across synapses, and produce our memories, thoughts and movements. Glial cells maintain homeostasis, produce myelin, and provide support and protection for the brain's neurons. Some glial cells (astrocytes) can even propagate intercellular calcium waves over long distances in response to stimulation, and release gliotransmitters in response to changes in calcium concentration. The extracellular matrix also provides support on the molecular level for the brain's cells.
At the organ level, the nervous system is composed of brain regions, such as the hippocampus in mammals or the mushroom bodies of the fruit fly.[5] These regions are often modular and serve a particular role within the general pathways of the nervous system. For example, the hippocampus is critical for forming memories. The nervous system also contains nerves, which are bundles of fibers that originate from the brain and spinal cord, and branch repeatedly to innervate every part of the body. Nerves are made primarily of the axons of neurons, along with a variety of membranes that wrap around and segregate them into nerve fascicles.
The vertebrate nervous system is divided into the central and peripheral nervous systems. The central nervous system (CNS) consists of the brain, retina, and spinal cord, while the peripheral nervous system (PNS) is made up of all the nerves outside of the CNS that connect it to the rest of the body. The PNS is further subdivided into the somatic and autonomic nervous systems. The somatic nervous system is made up of "afferent" neurons, which bring sensory information from the sense organs to the CNS, and "efferent" neurons, which carry motor instructions out to the muscles. The autonomic nervous system also has two subdivisions, the sympathetic and the parasympathetic, which are important for regulating the body's basic internal organ functions such as heartbeat, breathing, digestion, and salivation. Autonomic nerves, like somatic nerves, contain afferent and efferent fibers.
Orientation in neuroanatomy
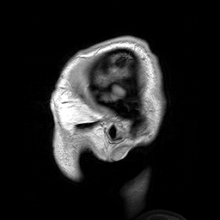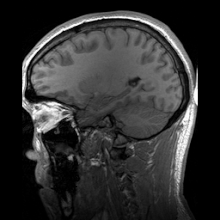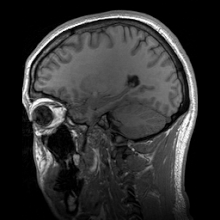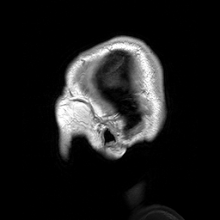
Para-sagittal MRI of the head in a patient with benign familial macrocephaly.
In anatomy in general and neuroanatomy in particular, several sets of topographic terms are used to denote orientation and location, which are generally referred to the body or brain axis (see Anatomical terms of location). The pairs of terms used most commonly in neuroanatomy are:
- Dorsal and ventral: dorsal loosely refers to the top or upper side, and ventral to the bottom or lower side. These descriptors originally referred to dorsum and ventrum – back and belly – of the body; the belly of most animals is oriented towards the ground; the erect posture of humans places our ventral aspect anteriorly, and the dorsal aspect becomes posterior. The case of the head and the brain is peculiar, since the belly does not properly extend into the head, unless we assume that the mouth represents an extended belly element. Therefore, in common use, those brain parts that lie close to the base of the cranium, and through it to the mouth cavity, are called ventral – i.e., at its bottom or lower side, as defined above – whereas dorsal parts are closer to the enclosing cranial vault.
- Rostral and caudal: rostral refers to the front of the body (towards the nose, or rostrum in Latin), and caudal to the tail end of the body (towards the tail; cauda in Latin). In Man, the directional terms "superior" and "inferior" essentially refer to this rostrocaudal dimension, because our body axis is roughly oriented vertically in the erect position. However, all vertebrates develop a kink in the neural tube that is still detectable in the adult central nervous system, known as the cephalic flexure. The latter bends the rostral part of the CNS at a 90 degree angle relative to the caudal part, at the transition between the forebrain and the brainstem and spinal cord. This change in axial dimension is problematic when trying to describe relative position and sectioning planes in the brain.
- Medial and lateral: medial refers to being close, or relatively closer, to the midline (the descriptor median means a position precisely at the midline. Lateral is the opposite (a position separated away from the midline)).
Note that such descriptors (dorsal/ventral, rostral/caudal; medial/lateral) are relative rather than absolute (e.g., a lateral structure may be said to lie medial to something else that lies even more laterally).
Commonly used terms for planes of orientation or planes of section in neuroanatomy are "sagittal", "transverse" or "coronal", and "axial" or "horizontal". Again in this case, the situation is different for swimming, creeping or quadrupedal (prone) animals than for Man, or other erect species, due to the changed position of the axis.
- A mid-sagittal plane divides the body and brain into left and right halves; sagittal sections in general are parallel to this median plane, moving along the medial-lateral dimension(see the image above). The term sagittal refers etymologically to the median suture between the right and left parietal bones of the cranium, known classically as sagittal suture, because it looks roughly like an arrow by its confluence with other sutures (sagitta; arrow in Latin).
- A section plane across any elongated form in principle is held to be transverse if it is orthogonal to the axis (e.g., a transverse section of a finger; if there is no length axis, there is no way to define such sections, or there are infinite possibilities). Therefore, transverse body sections in vertebrates are parallel to the ribs, which are orthogonal to the vertebral column, that represents the body axis both in animals and man. The brain also has an intrinsic longitudinal axis – that of the primordial elongated neural tube – which becomes largely vertical with the erect posture of Man, similarly as the body axis, except at its rostral end, as commented above. This explains that transverse spinal cord sections are roughly parallel to our ribs, or to the ground. However, this is only true for the spinal cord and the brainstem, since the forebrain end of the neural axis bends crook-like during early morphogenesis into the hypothalamus, where it ends; the orientation of true transverse sections accordingly changes, and is no longer parallel to the ribs and ground, but perpendicular to them; lack of awareness of this morphologic brain peculiarity (present in all vertebrate brains without exceptions) has caused and still causes erroneous thinking on forebrain brain parts. Acknowledging the singularity of rostral transverse sections, tradition has introduced a different descriptor for them, namely coronal sections. Coronal sections divide the forebrain from rostral (front) to caudal (back), forming a series orthogonal (transverse) to the local bent axis. The concept cannot be applied meaningfully to the brainstem and spinal cord, since there the coronal sections become horizontal to the axial dimension, being parallel to the axis.
- A coronal plane across the head and brain is modernly conceived to be parallel to the face (the etymology refers to corona or crown; the plane in which a king's crown sits on his head is not exactly parallel to the face, and exportation of the concept to less frontally endowed animals than us is obviously even more conflictive, but there is an implicit reference to the coronal suture of the cranium, which forms between the frontal and temporal/parietal bones, giving a sort of diadema configuration which is roughly parallel to the face). Coronal section planes thus essentially refer only to the head and brain, where a diadema makes sense, and not to the neck and body below.
- Horizontal sections by definition are aligned with the horizon. In swimming, creeping and quadrupedal animals the body axis itself is horizontal, and, thus, horizontal sections run along the length of the spinal cord, separating ventral from dorsal parts. Horizontal sections are orthogonal to both transverse and sagittal sections. Due to the axial bend in the brain (forebrain), true horizontal sections in that region are orthogonal to coronal (transverse) sections (as is the horizon relative to the face).
According to these considerations, the three directions of space are represented precisely by the sagittal, transverse and horizontal planes, whereas coronal sections can be transverse, oblique or horizontal, depending on how they relate to the brain axis and its incurvations.
Tools
Modern developments in neuroanatomy are directly correlated to the technologies used to perform research. Therefore, it is necessary to discuss the various tools that are available. Many of the histological techniques used to study other tissues can be applied to the nervous system as well. However, there are some techniques that have been developed especially for the study of neuroanatomy.
Cell staining
In biological systems, staining is a technique used to enhance the contrast of particular features in microscopic images.
Nissl staining uses aniline basic dyes to intensely stain the acidic polyribosomes in the rough endoplasmic reticulum, which is abundant in neurons. This allows researchers to distinguish between different cell types (such as neurons and glia), and neuronal shapes and sizes, in various regions of the nervous system cytoarchitecture.
The classic Golgi stain uses potassium dichromate and silver nitrate to fill selectively with a silver chromate precipitate a few neural cells (neurons or glia, but in principle any cells can react similarly). This so-called silver chromate impregnation procedure stains entirely or partially the cell bodies and neurites of some neurons -dendrites, axon- in brown and black, allowing researchers to trace their paths up to their thinnest terminal branches in a slice of nervous tissue, thanks to the transparency consequent to the lack of staining in the majority of surrounding cells. Modernly, Golgi-impregnated material has been adapted for electron-microscopic visualization of the unstained elements surrounding the stained processes and cell bodies, thus adding further resolutive power.
Histochemistry
Histochemistry uses knowledge about biochemical reaction properties of the chemical constituents of the brain (including notably enzymes) to apply selective methods of reaction to visualize where they occur in the brain and any functional or pathological changes. This applies importantly to molecules related to neurotransmitter production and metabolism, but applies likewise in many other directions chemoarchitecture, or chemical neuroanatomy.
Immunocytochemistry is a special case of histochemistry that uses selective antibodies against a variety of chemical epitopes of the nervous system to selectively stain particular cell types, axonal fascicles, neuropiles, glial processes or blood vessels, or specific intracytoplasmic or intranuclear proteins and other immunogenetic molecules, e.g., neurotransmitters. Immunoreacted transcription factor proteins reveal genomic readout in terms of translated protein. This immensely increases the capacity of researchers to distinguish between different cell types (such as neurons and glia) in various regions of the nervous system.
In situ hybridization uses synthetic RNA probes that attach (hybridize) selectively to complementary mRNA transcripts of DNA exons in the cytoplasm, to visualize genomic readout, that is, distinguish active gene expression, in terms of mRNA rather than protein. This allows identification histologically (in situ) of the cells involved in the production of genetically-coded molecules, which often represent differentiation or functional traits, as well as the molecular boundaries separating distinct brain domains or cell populations.
Genetically encoded markers
By expressing variable amounts of red, green, and blue fluorescent proteins in the brain, the so-called "brainbow" mutant mouse allows the combinatorial visualization of many different colors in neurons. This tags neurons with enough unique colors that they can often be distinguished from their neighbors with fluorescence microscopy, enabling researchers to map the local connections or mutual arrangement (tiling) between neurons.
Optogenetics uses transgenic constitutive and site-specific expression (normally in mice) of blocked markers that can be activated selectively by illumination with a light beam. This allows researchers to study axonal connectivity in the nervous system in a very discriminative way.
Non-invasive brain imaging
Magnetic resonance imaging has been used extensively to investigate brain structure and function non-invasively in healthy human subjects. An important example is diffusion tensor imaging, which relies on the restricted diffusion of water in tissue in order to produce axon images. In particular, water moves more quickly along the direction aligned with the axons, permitting the inference of their structure.
Viral-based methods
Certain viruses can replicate in brain cells and cross synapses. So, viruses modified to express markers (such as fluorescent proteins) can be used to trace connectivity between brain regions across multiple synapses.[6] Two tracer viruses which replicate and spread transneuronal/transsynaptic are the Herpes simplex virus type1 (HSV)[7] and the Rhabdoviruses.[8] Herpes simplex virus was used to trace the connections between the brain and the stomach, in order to examine the brain areas involved in viscero-sensory processing.[9] Another study injected herpes simplex virus into the eye, thus allowing the visualization of the optical pathway from the retina into the visual system.[10] An example of a tracer virus which replicates from the synapse to the soma is the pseudorabies virus.[11] By using pseudorabies viruses with different fluorescent reporters, dual infection models can parse complex synaptic architecture.[12]
Dye-based methods
Axonal transport methods use a variety of dyes (horseradish peroxidase variants, fluorescent or radioactive markers, lectins, dextrans) that are more or less avidly absorbed by neurons or their processes. These molecules are selectively transported anterogradely (from soma to axon terminals) or retrogradely (from axon terminals to soma), thus providing evidence of primary and collateral connections in the brain. These 'physiologic' methods (because properties of living, unlesioned cells are used) can be combined with other procedures, and have essentially superseded the earlier procedures studying degeneration of lesioned neurons or axons. Detailed synaptic connections can be determined by correlative electron microscopy.
Connectomics
Serial section electron microscopy has been extensively developed for use in studying nervous systems. For example, the first application of serial block-face scanning electron microscopy was on rodent cortical tissue.[13] Circuit reconstruction from data produced by this high-throughput method is challenging, and the Citizen science game EyeWire has been developed to aid research in that area.
Computational neuroanatomy
Is a field that utilizes various imaging modalities and computational techniques to model and quantify the spatiotemporal dynamics of neuroanatomical structures in both normal and clinical populations.
Model systems
Aside from the human brain, there are many other animals whose brains and nervous systems have received extensive study as model systems, including mice, zebrafish,[14]fruit fly,[15] and a species of roundworm called C. elegans. Each of these has its own advantages and disadvantages as a model system. For example, the C. elegans nervous system is extremely stereotyped from one individual worm to the next. This has allowed researchers using electron microscopy to map the paths and connections of all of the approximately 300 neurons in this species. The fruit fly is widely studied in part because its genetics is very well understood and easily manipulated. The mouse is used because, as a mammal, its brain is more similar in structure to our own (e.g., it has a six-layered cortex, yet its genes can be easily modified and its reproductive cycle is relatively fast).
Caenorhabditis elegans

Nervous system of a generic bilaterian animal, in the form of a nerve cord with segmental enlargements, and a "brain" at the front
The brain is small and simple in some species, such as the nematode worm, where the body plan is quite simple: a tube with a hollow gut cavity running from the mouth to the anus, and a nerve cord with an enlargement (a ganglion) for each body segment, with an especially large ganglion at the front, called the brain. The nematode Caenorhabditis elegans has been studied because of its importance in genetics.[16] In the early 1970s, Sydney Brenner chose it as a model system for studying the way that genes control development, including neuronal development. One advantage of working with this worm is that the nervous system of the hermaphrodite contains exactly 302 neurons, always in the same places, making identical synaptic connections in every worm.[17] Brenner's team sliced worms into thousands of ultrathin sections and photographed every section under an electron microscope, then visually matched fibers from section to section, to map out every neuron and synapse in the entire body, to give a complete connectome of the nematode.[18] Nothing approaching this level of detail is available for any other organism, and the information has been used to enable a multitude of studies that would not have been possible without it.[19]
Drosophila melanogaster
Drosophila melanogaster is a popular experimental animal because it is easily cultured en masse from the wild, has a short generation time, and mutant animals are readily obtainable.
Arthropods have a central brain with three divisions and large optical lobes behind each eye for visual processing. The brain of a fruit fly contains several million synapses, compared to at least 100 billion in the human brain. Approximately two-thirds of the Drosophila brain is dedicated to visual processing.
Thomas Hunt Morgan started to work with Drosophila in 1906, and this work earned him the 1933 Nobel Prize in Medicine for identifying chromosomes as the vector of inheritance for genes. Because of the large array of tools available for studying Drosophila genetics, they have been a natural subject for studying the role of genes in the nervous system.[20] The genome has been sequenced and published in 2000. About 75% of known human disease genes have a recognizable match in the genome of fruit flies. Drosophila is being used as a genetic model for several human neurological diseases including the neurodegenerative disorders Parkinson's, Huntington's, spinocerebellar ataxia and Alzheimer's disease. In spite of the large evolutionary distance between insects and mammals, many basic aspects of Drosophila neurogenetics have turned out to be relevant to humans. For instance, the first biological clock genes were identified by examining Drosophila mutants that showed disrupted daily activity cycles.[21]
See also
- Connectogram
- Outline of the human brain
- Outline of brain mapping
- List of regions in the human brain
- Medical image computing
- Neurology
- Neuroscience
- Computational anatomy
References
^ Atta, H. M. (1999). "Edwin Smith Surgical Papyrus: The Oldest Known Surgical Treatise". American Surgeon. 65 (12): 1190–1192..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ ab Rose, F (2009). "Cerebral Localization in Antiquity". Journal of the History of the Neurosciences. 18 (3): 239–247. doi:10.1080/09647040802025052.
^ Ginn, S. R.; Lorusso, L. (2008). "Brain, Mind, and Body: Interactions with Art in Renaissance Italy". Journal of the History of the Neurosciences. 17 (3): 295–313. doi:10.1080/09647040701575900.
^ Neher, A (2009). "Christopher Wren, Thomas Willis and the Depiction of the Brain and Nerves". Journal of Medical Humanities. 30 (3): 191–200. doi:10.1007/s10912-009-9085-5.
^ Mushroom Bodies of the Fruit Fly Archived 2012-07-16 at Archive.today
^ Ginger, M.; Haberl, M.; Conzelmann, K.-K.; Schwarz, M.; Frick, A. (2013). "Revealing the secrets of neuronal circuits with recombinant rabies virus technology". Front. Neural Circuits. 7. doi:10.3389/fncir.2013.00002.
^ McGovern, AE; Davis-Poynter, N; Rakoczy, J; Phipps, S; Simmons, DG; Mazzone, SB (2012). "Anterograde neuronal circuit tracing using a genetically modified herpes simplex virus expressing EGFP". J Neurosci Methods. 209 (1): 158–67. doi:10.1016/j.jneumeth.2012.05.035. PMID 22687938.
^ Kuypers HG, Ugolini G (February 1990). "Viruses as transneuronal tracers". Trends in Neurosciences. 13 (2): 71–5. doi:10.1016/0166-2236(90)90071-H. PMID 1690933.
^ Rinaman L, Schwartz G (March 2004). "Anterograde transneuronal viral tracing of central viscerosensory pathways in rats". The Journal of Neuroscience. 24 (11): 2782–6. doi:10.1523/JNEUROSCI.5329-03.2004. PMID 15028771.
^ Norgren RB, McLean JH, Bubel HC, Wander A, Bernstein DI, Lehman MN (March 1992). "Anterograde transport of HSV-1 and HSV-2 in the visual system". Brain Research Bulletin. 28 (3): 393–9. doi:10.1016/0361-9230(92)90038-Y. PMID 1317240.
^ Card, J. P. (2001). "Pseudorabies virus neuroinvasiveness: A window into the functional organization of the brain". Advances in Virus Research.
^ Card, J. P. (2011). "A Dual Infection Pseudorabies Virus Conditional Reporter Approach to Identify Projections to Collateralized Neurons in Complex Neural Circuits". PLoS ONE. 6: e21141. doi:10.1371/journal.pone.0021141. PMC 3116869. PMID 21698154.
^ Denk, W; Horstmann, H (2004). "Serial Block-Face Scanning Electron Microscopy to Reconstruct Three-Dimensional Tissue Nanostructure". PLoS Biology. 2: e329. doi:10.1371/journal.pbio.0020329. PMC 524270. PMID 15514700.
^ Wullimann, Mario F.; Rupp, Barbar; Reichert, Heinrich (1996). Neuroanatomy of the zebrafish brain: a topological atlas. ISBN 3-7643-5120-9.
^ Atlas of the Drosophila Brain
^ "WormBook: The online review of C. elegans biology". Retrieved 2011-10-14.
^ Hobert, Oliver (2005). The C. elegans Research Community, ed. "Specification of the nervous system". WormBook: 1–19. doi:10.1895/wormbook.1.12.1. PMC 4781215. PMID 18050401.
^ White, JG; Southgate, E; Thomson, JN; Brenner, S (1986). "The Structure of the Nervous System of the Nematode Caenorhabditis elegans". Philosophical Transactions of the Royal Society B. 314 (1165): 1–340. doi:10.1098/rstb.1986.0056. PMID 22462104.
^ Hodgkin, J (2001). "Caenorhabditis elegans". In Brenner S, Miller JH. Encyclopedia of Genetics. Elsevier. pp. 251–256. ISBN 978-0-12-227080-2.CS1 maint: Uses editors parameter (link)
^ "Flybrain: An online atlas and database of the drosophila nervous system". Archived from the original on 2016-05-16. Retrieved 2011-10-14.
^ Konopka, RJ; Benzer, S (1971). "Clock Mutants of Drosophila melanogaster". Proc. Natl. Acad. Sci. U.S.A. 68 (9): 2112–6. doi:10.1073/pnas.68.9.2112. PMC 389363. PMID 5002428.
External links
| Wikimedia Commons has media related to Neuroanatomy. |
Neuroanatomy, an annual journal of clinical neuroanatomy- Mouse, Rat, Primate and Human Brain Atlases (UCLA Center for Computational Biology)
- brainmaps.org: High-Resolution Neuroanatomically-Annotated Brain Atlases
- BrainInfo for Neuroanatomy
Brain Gene Expression Map, mouse gene expression neuroanatomical resource from St. Jude Children's Research Hospital
Brain Architecture Management System, several atlases of brain anatomy
White Matter Atlas, Diffusion Tensor Imaging Atlas of the Brain's White Matter Tracts